Mate-pair signatures for inversion style mis-assemblies. (a) Two copy,... | Download Scientific Diagram

Ion Torrent on Twitter: "Ion TrueMate Library Kit technology generates mate- pair libraries with insert size of 2-8kb http://t.co/dkES1GZZv2 http://t.co/KpUjTRi2Ea" / Twitter

Overlapping Chimeric paired-end alignments are excluded (e.g. in FFPE data) · Issue #285 · alexdobin/STAR · GitHub

Structural variant detection in cancer genomes: computational challenges and perspectives for precision oncology | npj Precision Oncology

ChimPipe – Accurate detection of fusion genes and transcription-induced chimeras from RNA-seq data | RNA-Seq Blog

How chimeric reads result from ERV integration. (A to D) A guide to... | Download Scientific Diagram

HiTea: a computational pipeline to identify non-reference transposable element insertions in Hi-C data | bioRxiv
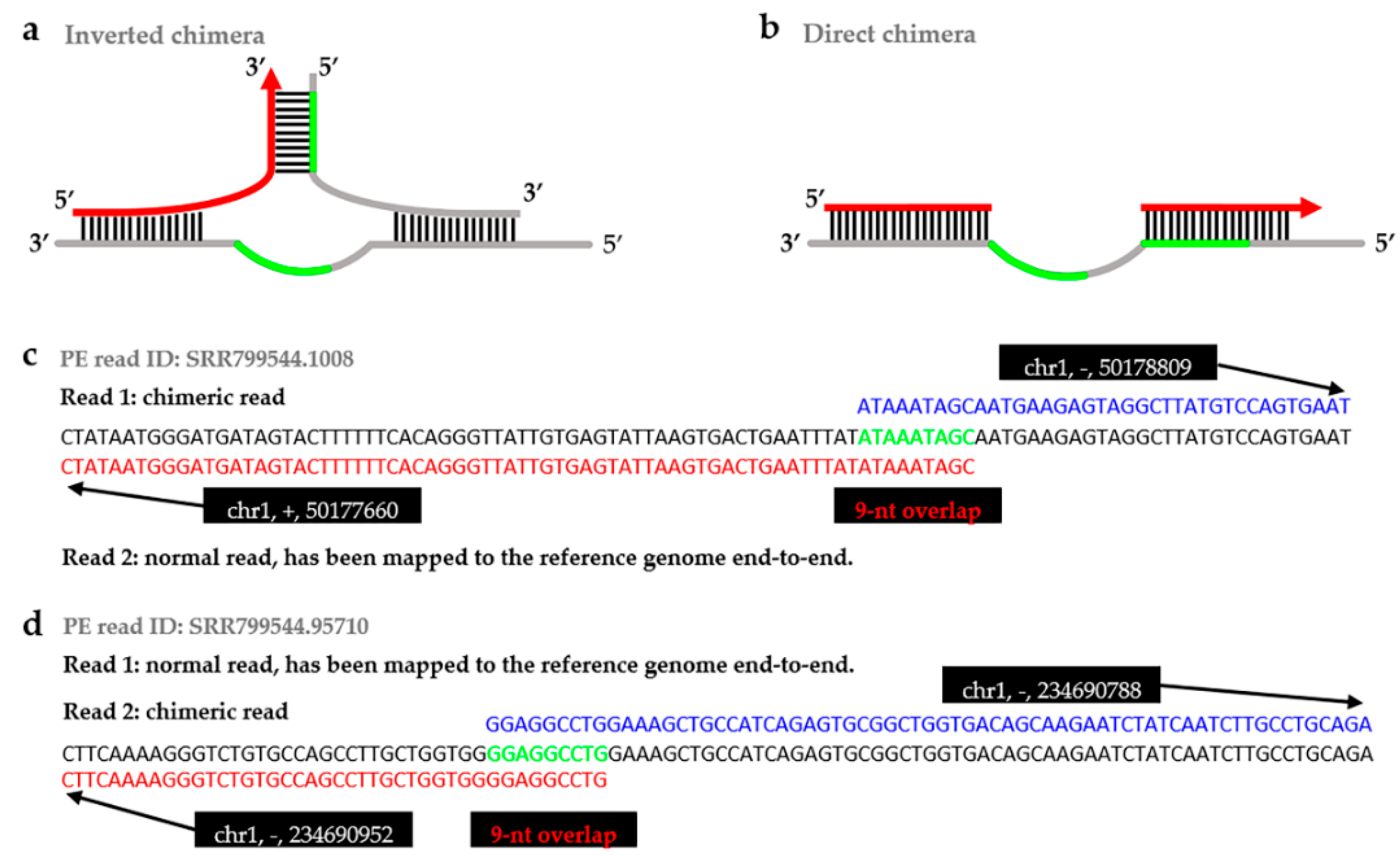
IJMS | Free Full-Text | ChimeraMiner: An Improved Chimeric Read Detection Pipeline and Its Application in Single Cell Sequencing